Challenges in the Study of Biological Membranes using Molecular Dynamics Simulations
DOI:
https://doi.org/10.36790/epistemus.v19i38.357Keywords:
lipids, bilayer, simulations, membrane, phasesAbstract
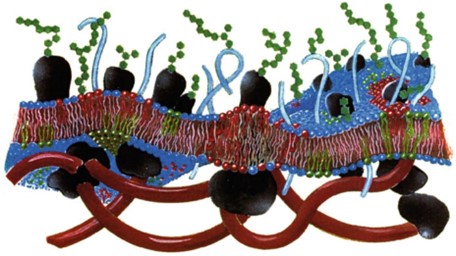
In the study of model membrane simulations, the selection of components is pivotal. Even when considering only the bilayer, the essential core of the plasma membrane, different lipids generate various interactions, influencing system’s behavior and giving rise to diverse biological processes. Accounting for appropriate temporal scales is essential, as distinct biological phenomena operate within specific time intervals. Choosing an inappropriate scale might overlook intrinsic details of the phenomenon under investigation.
In the present study, we delve into two simulation scales: the detailed resolution of all atoms and the coarse-grained simplification, emphasizing their impact on accuracy and computational performance. We address complexity by studying individual components, such as the bilayer. This approach provides a valuable perspective for comprehending intricate biological processes in the plasma membrane, emphasizing the significance of careful choices in in the simulation of biomimetic systems.
Downloads
References
M. Feig, G. Nawrocki, I. Yu, “Challenges and opportunities in connecting simulations with experiment via molecular dynamics of cellular environments”, J. Phys. Conf. Ser, vol. 1036, no. 1, Enero 2018, DOI: http://10.1088/1742-6596/1036/1/012010. DOI: https://doi.org/10.1088/1742-6596/1036/1/012010
J. D. Murray, J. J. Lange, H. Bennett-Lenane, R. Holm, “Advancing algorithmic drug product development: Recommendations for machine learning approaches in drug formulation”, Eur. J. Pharm. Sci., vol. 191, Diciembre 2023, DOI: http://10.1016/j.ejps.2023.106562. DOI: https://doi.org/10.1016/j.ejps.2023.106562
I. Vattulainen, T. Rog, “Lipid simulations: A perspective on lipids in action”, Cold Spring Harb. Perspect. Biol., vol. 3, no. 4, Abril 2011, DOI: http://10.1101/cshperspect.a004655 . DOI: https://doi.org/10.1101/cshperspect.a004655
T. Heimburg, R. L. Biltonen, “A Monte Carlo simulation study of protein-induced heat capacity changes and lipid-induced protein clustering”, Biophys. J., vol. 70, pp. 84-96, Enero 1996, DOI: http://10.1016/S0006-3495(96)79551-6. DOI: https://doi.org/10.1016/S0006-3495(96)79551-6
R. X . Gu, S. Baoukina, D. P. Tieleman, “Phase separation in atomistic simulations of model membranes” J. Am. Chem. Soc., vol. 142, no. 6, pp. 2844-2856, Enero 2020, DOI: http://10.1021/jacs.9b11057. DOI: https://doi.org/10.1021/jacs.9b11057
L. Karami, “Interactions of neutral and protonated Tamoxifen with the DPPC lipid bilayer using molecular dynamics simulation”, Steroids, vol. 194, Enero 2023, DOI: http://10.1016/j.steroids.2023.109225. DOI: https://doi.org/10.1016/j.steroids.2023.109225
H. I. Ingólfsson, M. N. Melo, “Lipid Organization of the plasma membrane”, Journal of the American Chemical Society, vol.136, no. 41, pp. 14554-14559, Septiembre 2014, DOI: http://10.1021/ja507832e. DOI: https://doi.org/10.1021/ja507832e
E. Sezgin, “Giant plasma membrane vesicles to study plasma membrane structure and dynamics”, Biochim. Biophys. Acta. Biomembr., vol. 1864, no. 4, Abril 2022, DOI: http://10.1016/j.bbamem.2021.183857. DOI: https://doi.org/10.1016/j.bbamem.2021.183857
J. D. Nickels, “Lipid Rafts: Buffers of Cell Membrane Physical Properties”, J. Phys. Chem B, vol. 123, pp. 2050-2056, Enero 2019, DOI: https://doi.org/10.1021/acs.jpcb.8b12126. DOI: https://doi.org/10.1021/acs.jpcb.8b12126
E. Sezgin, I. Levental, S. Mayor, C. Eggeling, “The mystery of membrane organization: Composition, regulation and roles of lipid rafts”, Nature Reviews Molecular Cell Biology, vol. 18, no. 6, pp. 361–374, Junio 2017, DOI: https://doi.org/10.1038/nrm.2017.16. DOI: https://doi.org/10.1038/nrm.2017.16
S. L. Veacht, N. Rogers, A. Decker, S. A. Shelby, “The plasma membranes as an adaptable fluid mosaic”, Biochim. Biophys. Acta Biomembr., vol. 1865, no. 3, Marzo 2023, DOI: https://doi.org/10.1016/j.bbamem.2022.184114. DOI: https://doi.org/10.1016/j.bbamem.2022.184114
M. Doktorova, G. Khelashvili, R. Ashkar, M. F. Brown, “Molecular simulations and NMR reveal how lipid fluctuations affect membrane mechanics”, Biophys. J., vol. 122, no. 6, pp. 984-1002, Diciembre 2022, DOI: https://doi.org/10.1016/j.bpj.2022.12.007. DOI: https://doi.org/10.1016/j.bpj.2022.12.007
S. Qian, V. K. Sharma, L. A. Clifton, “Understanding the structure and dynamics of complex biomembrane interactions by neutron scattering techniques”, Langmuir, vol. 36, pp. 15189-15211, Diciembre 2020, DOI: https://doi.org/10.1021/acs.langmuir.0c02516. DOI: https://doi.org/10.1021/acs.langmuir.0c02516
R. Metzler, J. H. Jeon, A.G. Cherstvy, “Non-brownian diffusion in lipid membranes: experiments and simulations”, Biochim. Biophys. Acta. Biomembr., vol. 1858, no. 10, pp. 2451-2467, Octubre 2016, DOI: https://doi.org/10.1016/j.bbamem.2016.01.022. DOI: https://doi.org/10.1016/j.bbamem.2016.01.022
L. Porcar, Y. Gerelli, “On the lipid flip-flop and phase transition coupling”, Soft Matter, vol. 16, no. 33, pp. 7696-7703, Agosto 2020, DOI: https://doi.org/10.1039/D0SM01161D. DOI: https://doi.org/10.1039/D0SM01161D
E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, “CHARMM-GUI membrane builder toward realistic biological membrane simulations”, J. Comput. Chem., vol.35, no. 27, pp. 1997-2004, Octubre 2014, DOI: https://doi.org/10.1002/jcc.23702. DOI: https://doi.org/10.1002/jcc.23702
S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman, “The MARTINI force field: Coarse grained model for biomolecular simulations”, J. Phys. Chem. B., vol. 111, pp. 7812-7824, Junio 2007, DOI: https://doi.org/10.1021/jp071097f. DOI: https://doi.org/10.1021/jp071097f
H. Rakhshani, E. Dehghanian, A. Rahati, “Enhanced GROMACS: toward a better numerical simulation framework”, J. Mol. Model, vol. 25, no. 12, Noviembre 2019, DOI: https://doi.org/10.1007/s00894-019-4232-z. DOI: https://doi.org/10.1007/s00894-019-4232-z
P. Van Der Ploeg, H. J. C. Berendsen, “Molecular dynamics of a bilayer membrane”, J. Chem. Phys, vol. 76, no. 6, pp. 3271-3276, Marzo 1982, DOI: https://doi.org/10.1007/978-1-62703-017-5_15. DOI: https://doi.org/10.1063/1.443321
S. J. Marrink, V. Corradi, H. I. Ingólfsson, D. P. Tieleman, “Computational Modelling of realistic cell membranes”, Chem. Rev., vol. 119, no. 9, pp. 6184-6226, Mayo 2019, DOI: https://doi.org/10.1021/acs.chemrev.8b00460. DOI: https://doi.org/10.1021/acs.chemrev.8b00460
J. Risbo, M. M. Sperotto, O. G. Mouritsen, “Theory of phase equilibria and critical mixing points in binary lipid bilayers”, J. Chem. Phys, vol. 103, no. 9, pp. 3643-3656, Mayo 1995, DOI: https://doi.org/10.1063/1.470041. DOI: https://doi.org/10.1063/1.470041
O. G. Mouritsen, L. A. Bagatolli, Life - as a matter of fat lipids in a membrane biophysics perspective. Segunda edición. Suiza: Springer International Publishing Switzerland, 2005, DOI: https://doi.org/10.13140/RG.2.1.1190.4723.
D. Marsh, “Cholesterol-induced fluid membrane domains: A compendium of lipid-raft ternary phase diagrams”, Biochimica et Biophysica Acta – Biomembranes, vol. 1788, no. 10, pp. 2114–2123, Agosto 2009, DOI: https://doi.org/10.1016/j.bbamem.2009.08.004. DOI: https://doi.org/10.1016/j.bbamem.2009.08.004
I. Levental, K. R. Levental, F. A. Heberle, “Lipid Rafts: Controversies Resolved, Mysteries Remain”, Trends in Cell Biology, vol. 30, no. 5, pp. 341–353, Mayo 2020, DOI: https://doi.org/10.1016/j.tcb.2020.01.009. DOI: https://doi.org/10.1016/j.tcb.2020.01.009
K. Simons, E. Ikonen, “Functional rafts in cell membranes”, Nature, vol. 387, pp. 569–572, Junio 1997, DOI: https://doi.org/10.1038/42408. DOI: https://doi.org/10.1038/42408
I. Levental, S. L. Veatch, “The Continuing Mystery of Lipid Rafts”, Journal of Molecular Biology, vol. 428, no. 24, pp. 4749–4764, Agosto 2016, DOI: https://doi.org/10.1016/j.jmb.2016.08.022. DOI: https://doi.org/10.1016/j.jmb.2016.08.022
A. D. Maldonado Arce, C. Contreras Aburto, F. F. Rosales, J. A. Arvayo Zatarain, E. Urrutia Bañuelos, “Métodos de simulación computacional en biología”, EPISTEMUS, no. 21, 84-92, Diciembre 2016, DOI: https://doi.org/10.36790/epistemus.v10i21.38. DOI: https://doi.org/10.36790/epistemus.v10i21.38
M. P. Allen, D. J. Tildesley, Computer Simulation of Liquids, 2nd. Ed., Oxford University Press, 2017, DOI: https://doi.org/10.1093/oso/9780198803195.001.0001. DOI: https://doi.org/10.1093/oso/9780198803195.001.0001
D. Frenkel, B. Smit, Understanding Molecular Simulation From Algorithms to Applications, 2nd. Ed., Academic Press, 2002, DOI: https://doi.org/10.1063/1.881812. DOI: https://doi.org/10.1063/1.881812
M. Parrinello, A. Rahman, “Polymorphic transitions in single crystals: A new molecular dynamics method”, J. Appl. Phys., vol. 52, no. 12, pp. 7182-7190, Diciembre 1981, DOI: https://doi.org/10.1063/1.328693. DOI: https://doi.org/10.1063/1.328693
S. Nose, “A unified formulation of the constant temperature molecular dynamics methods”, J. Phys. Chem. Lett., vol. 81, no. 1, pp. 511-519, Julio 1984, DOI: https://doi.org/10.1063/1.447334. DOI: https://doi.org/10.1063/1.447334
R. W. Pastor, A. D. MacKerell, “Development of the CHARMM force field for lipids”, J. Phys. Chem. Lett., vol. 2, no. 13, pp. 1526-1532, Junio 2011, DOI: https://doi.org/10.1021/jz200167q. DOI: https://doi.org/10.1021/jz200167q
P. S. Nerenberg, T. Head-Gordon, “New developments in force fields for biomolecular simulations”, Current Opinion in Structural Biology, vol. 49, pp. 129-138, Abril 2018, DOI: https://doi.org/10.1016/j.sbi.2018.02.002. DOI: https://doi.org/10.1016/j.sbi.2018.02.002
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 EPISTEMUS

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.
The magazine acquires the patrimonial rights of the articles only for diffusion without any purpose of profit, without diminishing the own rights of authorship.
The authors are the legitimate owners of the intellectual property rights of their respective articles, and in such quality, by sending their texts they express their desire to collaborate with the Epistemus Magazine, published biannually by the University of Sonora.
Therefore, freely, voluntarily and free of charge, once accepted the article for publication, they give their rights to the University of Sonora for the University of Sonora to edit, publish, distribute and make available through intranets, Internet or CD said work, without any limitation of form or time, as long as it is non-profit and with the express obligation to respect and mention the credit that corresponds to the authors in any use that is made of it.
It is understood that this authorization is not an assignment or transmission of any of your economic rights in favor of the said institution. The University of Sonora guarantees the right to reproduce the contribution by any means in which you are the author, subject to the credit being granted corresponding to the original publication of the contribution in Epistemus.
Unless otherwise indicated, all the contents of the electronic edition are distributed under a license for use and Creative Commons — Attribution-NonCommercial-ShareAlike 4.0 International — (CC BY-NC-SA 4.0) You can consult here the informative version and the legal text of the license. This circumstance must be expressly stated in this way when necessary.
The names and email addresses entered in this journal will be used exclusively for the purposes established in it and will not be provided to third parties or for their use for other purposes.